R is a langage that has been created to manipulate data. This means that R has a lot of built-in tools and functions such as data frames, vectors, matrices and decision trees out of the box to cover your basic needs of data science and even machine learning.
Considering this, it might be tempting to not look further and use the native options provided, but this is a mistake: the alternatives procures a lot of advantages and little to no disadvantages.
They are both faster, and easier to write, and are also more customizable. In this article, we’ll see 4 domains of data science where base R is outmatched by the alternatives.
Data manipulation
Data manipulation in base R is possible with no additional libraries, but it’s slower and less powerful than the alternatives proposed here. Here is the dataset used in the following examples :

Base R
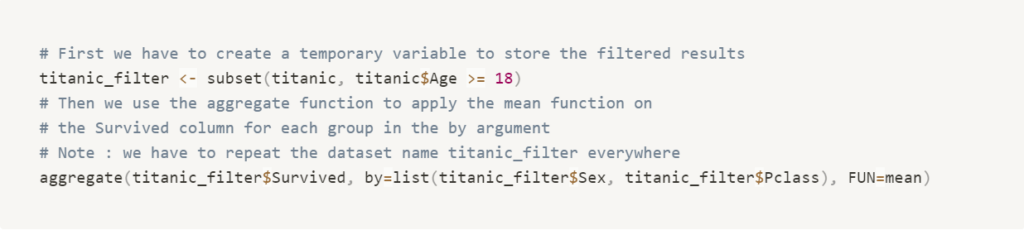
data.table
The fastest library for data manipulation in R, with a concise syntax, that can also read and write files. It can modify the data frame directly by reference to avoid data copy, so you can work on large in memory datasets. It has no dependencies so it’s very quick to install.

dplyr
Another alternative to base R, it has a verbose (and perhaps more readable) syntax. It considers data as immutable to avoid unwanted side effects at the cost of processing time. It’s still much faster at processing data than base R, but slower than data.table. It comes with a lot of dependencies from the tidyverse, it can take a while to install, but you’ll have many others useful libraries.

Machine Learning
The ML environment in R is very different from Python: In R, most of the time, a library contains only one algorithm, and each algorithm can be implemented in different ways, in different librairies.
For example, there are at least 8 libraries implementing a Random Forest, and 12 more implements variations of decision trees. It can be difficult to choose the best one for you, especially because they all have different methods and arguments and testing them all is not a simple process.
This makes it more complicated to try different algorithms on the same dataset.
H2O
H2O is a ML library written in Java with interfaces in R, Python, and many others languages. It comes with around 20 algorithms such as Random Forest, GBM, Deep Neural Networks, K-Means, PCA and more.
It handles natively missing data, categorical columns, cross validation, multithreading and it computes all necessary metrics at the end of the model training. They are accessible from R and from a local web page to directly view all kinds of interactive graphics.
The main benefit of having multiple algorithms in a single library is the unified API to call them. It’s very easy to try different models on the same dataset as shown in the following example.
Data prep

Train 3 differents models
Thanks to having the same API, it’s very easy to train multiples models on the same dataset with the same arguments. Each algorithm has specific parameters, but usually the defaults gives pretty good results.

Data Visualization
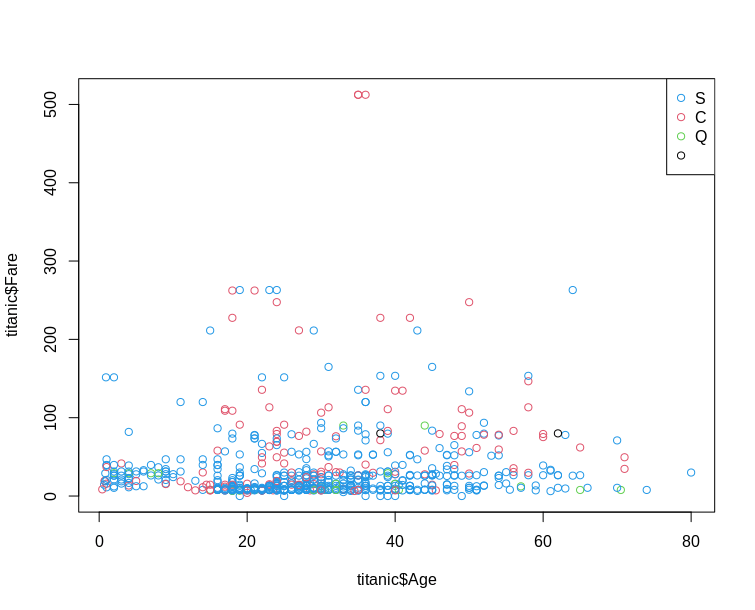
Base R already contains an easy to use plot method, but the output is basic and not very customizable. The main advantage is that it works on a lot of different objects and the syntax is easy to learn.
Base R

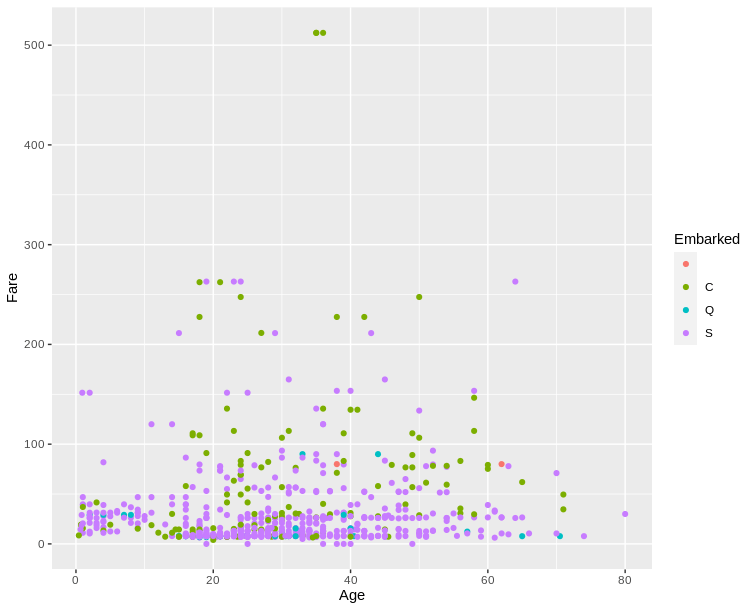
ggplot2
ggplot is harder to learn when you are beginning R because of its specific syntax, but it is much easier to customize and in general the graphs produced are more readable.
Once you get used to the syntax, it becomes much faster to use than base R. The graphics produced tends to be cleaner and more readable.

Data Storage
It’s often useful to store datasets, model metrics, and other information to disk to save time and keep track of past executions. There are multiple ways to do it, each with advantages and disavantages.
Base R
There are 2 native solutions to store data to disk. Either write a data frame as csv, or store any object as binary in a R specific format. Storing a .csv makes it easy to read from any other application or langage, but the file takes up more space, and it only works on tabular data. The binary storage is faster but it’s specific to R and cannot be used to share data to others applications. For each example, we’ll see how to write and then read the dataset.

Data.table
If you need to store data as .csv, data.table is the fastest way to do it. It’s tens to hundreds of times faster than base R, and you get the same file at the end.
arrow
If you need speed, a small file size, and a standard format, the arrow library combines all 3 when storing data as a parquet file. It’s not quite as fast as data.table but having a parquet file can be very useful. For example you can store the file on an hdfs or aws s3, and directly declare a Impala / Athena table on the file itself, no additional copy required. To makes things even easier, the arrow library accepts s3 URI when reading and writing data.










